Calculate Geospatial Weighted Averages from Monthly Time Series#
Authors:
Date: 05/27/22
Related APIs:
The data used in this example can be found through the Earth System Grid Federation (ESGF) search portal.
Overview#
A common data reduction in geophysical sciences is to produce spatial averages. Spatial averaging functionality in xcdat allows users to quickly produce area-weighted spatial averages for selected regions (or full dataset domains).
In the example below, we demonstrate the opening of a (remote) dataset and spatial averaging over the global, tropical, and Niño 3.4 domains.
Notebook Setup#
Create an Anaconda environment for this notebook using the command below, then select the kernel in Jupyter.
conda create -n xcdat_notebook -c conda-forge python xarray netcdf4 xcdat xesmf matplotlib nc-time-axis jupyter
xesmfis required for horizontal regridding with xESMFmatplotlibis an optional dependency required for plotting with xarraync-time-axisis an optional dependency required formatplotlibto plotcftimecoordinates
[1]:
%matplotlib inline
import matplotlib.pyplot as plt
import xcdat
1. Open the Dataset#
We are using xarray’s OPeNDAP support to read a netCDF4 dataset file directly from its source. The data is not loaded over the network until we perform operations on it (e.g., temperature unit adjustment).
More information on the xarray’s OPeNDAP support can be found here.
[2]:
filepath = "https://esgf-data1.llnl.gov/thredds/dodsC/css03_data/CMIP6/CMIP/CSIRO/ACCESS-ESM1-5/historical/r10i1p1f1/Amon/tas/gn/v20200605/tas_Amon_ACCESS-ESM1-5_historical_r10i1p1f1_gn_185001-201412.nc"
ds = xcdat.open_dataset(filepath)
# Unit adjust (-273.15, K to C)
ds["tas"] = ds.tas - 273.15
ds
[2]:
<xarray.Dataset>
Dimensions: (time: 1980, bnds: 2, lat: 145, lon: 192)
Coordinates:
* time (time) datetime64[ns] 1850-01-16T12:00:00 ... 2014-12-16T12:00:00
* lat (lat) float64 -90.0 -88.75 -87.5 -86.25 ... 86.25 87.5 88.75 90.0
* lon (lon) float64 0.0 1.875 3.75 5.625 ... 352.5 354.4 356.2 358.1
height float64 2.0
Dimensions without coordinates: bnds
Data variables:
time_bnds (time, bnds) datetime64[ns] 1850-01-01 1850-02-01 ... 2015-01-01
lat_bnds (lat, bnds) float64 -90.0 -89.38 -89.38 ... 89.38 89.38 90.0
lon_bnds (lon, bnds) float64 -0.9375 0.9375 0.9375 ... 357.2 357.2 359.1
tas (time, lat, lon) float32 -27.19 -27.19 -27.19 ... -25.29 -25.29
Attributes: (12/48)
Conventions: CF-1.7 CMIP-6.2
activity_id: CMIP
branch_method: standard
branch_time_in_child: 0.0
branch_time_in_parent: 87658.0
creation_date: 2020-06-05T04:06:11Z
... ...
variant_label: r10i1p1f1
version: v20200605
license: CMIP6 model data produced by CSIRO is li...
cmor_version: 3.4.0
tracking_id: hdl:21.14100/af78ae5e-f3a6-4e99-8cfe-5f2...
DODS_EXTRA.Unlimited_Dimension: time2. Global#
[3]:
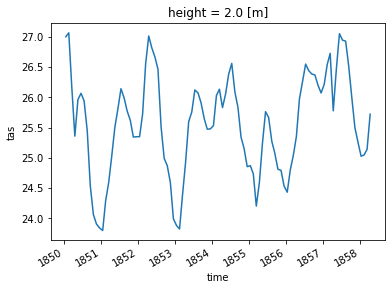
ds_global_avg = ds.spatial.average("tas")
[4]:
ds_global_avg.tas
[4]:
<xarray.DataArray 'tas' (time: 1980)>
array([12.52127071, 13.09115223, 13.60703132, ..., 15.5767848 ,
14.65664621, 13.84951678])
Coordinates:
* time (time) datetime64[ns] 1850-01-16T12:00:00 ... 2014-12-16T12:00:00
height float64 2.0[5]:
# Plot the first 100 time steps
ds_global_avg.tas.isel(time=slice(0, 100)).plot()
[5]:
[<matplotlib.lines.Line2D at 0x7fac9d6aee80>]
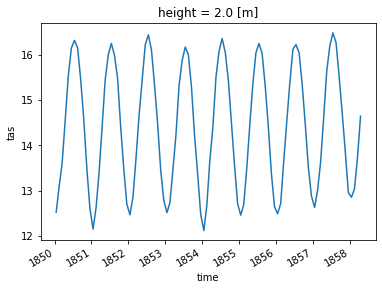
3. Tropical Region#
[6]:
ds_trop_avg = ds.spatial.average("tas", lat_bounds=(-25, 25))
[7]:
ds_trop_avg.tas
[7]:
<xarray.DataArray 'tas' (time: 1980)>
array([25.24722608, 25.61795924, 25.96516235, ..., 26.79536823,
26.67771602, 26.27182383])
Coordinates:
* time (time) datetime64[ns] 1850-01-16T12:00:00 ... 2014-12-16T12:00:00
height float64 2.0[8]:
# Plot the first 100 time steps
ds_trop_avg.tas.isel(time=slice(0, 100)).plot()
[8]:
[<matplotlib.lines.Line2D at 0x7fac9d58ff70>]
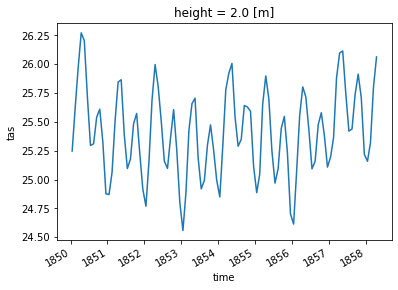
4. Nino 3.4 Region#
Niño 3.4 (5N-5S, 170W-120W): The Niño 3.4 anomalies may be thought of as representing the average > equatorial SSTs across the Pacific from about the dateline to the South American coast. The Niño > 3.4 index typically uses a 5-month running mean, and El Niño or La Niña events are defined when > the Niño 3.4 SSTs exceed +/- 0.4C for a period of six months or more.”
—https://climatedataguide.ucar.edu/climate-data/nino-sst-indices-nino-12-3-34-4-oni-and-tni
[9]:
ds_nino_avg = ds.spatial.average("tas", lat_bounds=(-5, 5), lon_bounds=(190, 240))
[10]:
ds_nino_avg.tas
[10]:
<xarray.DataArray 'tas' (time: 1980)>
array([27.00284678, 27.06796429, 26.18095324, ..., 27.17515272,
27.30917002, 27.38399379])
Coordinates:
* time (time) datetime64[ns] 1850-01-16T12:00:00 ... 2014-12-16T12:00:00
height float64 2.0[11]:
# Plot the first 100 time steps
ds_nino_avg.tas.isel(time=slice(0, 100)).plot()
[11]:
[<matplotlib.lines.Line2D at 0x7fac9d515eb0>]